Patients with a neurodegenerative disease, such as Alzheimer’s disease, epilepsy, or schizophrenia, are prone to showing increased rates of hippocampal atrophy. Monitoring hippocampal volume changes by segmenting the structure on MRI scans can, therefore, support the tracking of disease progression and the evaluation of drug efficiency in clinical trials.
Tracking hippocampal shape changes and volume loss have piqued the interest of professionals involved in the field of radiotherapy for brain tumor treatment. Patients who receive radiotherapy often suffer from cognitive decline after their treatment, which possibly relates to decreased volumes of the hippocampi following exposure to radiation. To investigate the correlation between radiation dose and hippocampal atrophy, it is crucial to determine hippocampal volume.
Determining hippocampal volume manually: a time-consuming challenge
Unfortunately, segmenting the hippocampus manually on a brain MRI scan is a rather tedious task. Delineating the structure on multiple slices can take up a good portion of the radiologists’ precious time. Current image acquisition techniques are not perfect. Technological constraints of an MRI system lead to images with limited resolution and suboptimal contrast between different structures. Especially on MRI images of the brain, it is often hard to distinguish smaller structures. Additionally, the image intensities of the different structures are similar - complicating the segmentation process. Manually segmenting structures on a medical image is hardly a straightforward exercise. Even trained professionals, such as radiologists, can segment the same structures with highly different results. In extreme cases, this might even lead to different insights supporting different diagnoses or treatment decisions. It speaks for itself that using manual segmentation approaches for such investigations has its flaws.
Figure 1: Manual hippocampus segmentation can be a time consuming task.
Can't we determine the hippocampal volume automatically?
Luckily for us, we can automate the process, which is a great way of obtaining valuable hippocampal segmentations without wasting too much time. There are a number of algorithms available that can segment hippocampal volume on MRI scans. Below, we discuss a few examples.
Fully automated hippocampal segmentation
FreeSurfer and FSL-FIRST (for the detail-oriented ones amongst us: this stands for FMRIB Software Library - FMRIB's Integrated Registration and Segmentation Tool) are two well-known examples of fully automated methods for hippocampus segmentation. FreeSurfer uses deformable atlas-based segmentation methods, which compare the input image with one or more atlases and match each mapped voxel to the right anatomical structure. FSL-FIRST is a surface-based shape segmentation method. Surface-based methods construct a triangulated mesh on the surface of the structure to be segmented. Through stepwise adjustments, this mesh gets optimized to fit the surface of the structure better and better, eventually resulting in a structure segmentation.
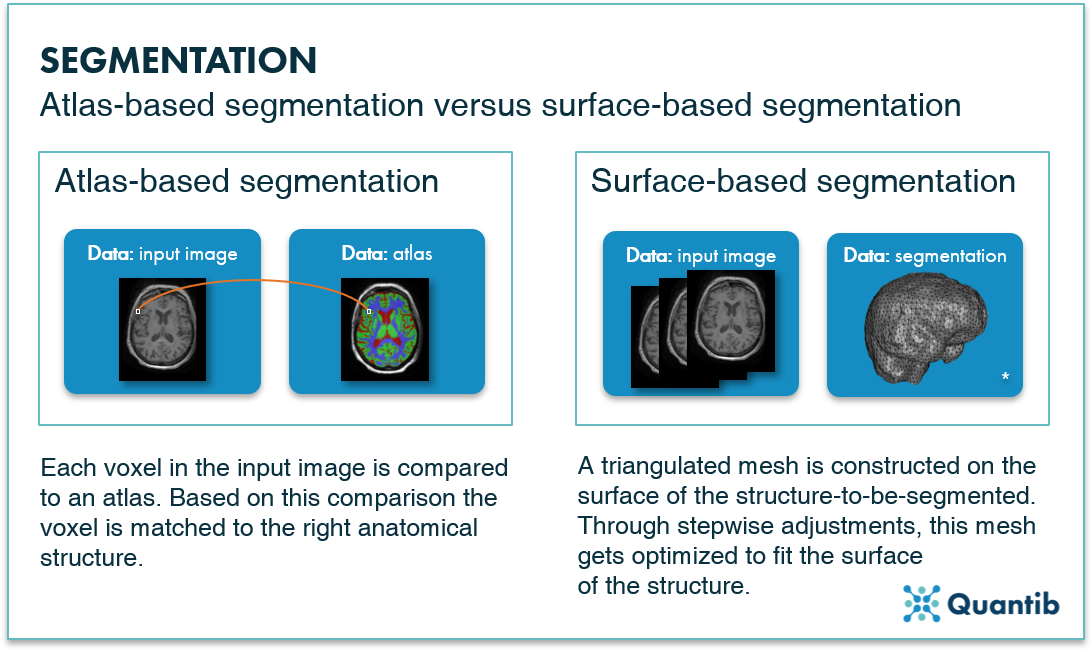
Figure 2: Atlas-based segmentation methods perform a voxel-wise comparison of input image with an atlas. Surface-based segmentation methods construct a triangulated mesh around a structure. * Brain mesh image adopted from Lobos et al.1
There are numerous studies comparing these two methods to manual segmentations and the general conclusion is that, although the results look very promising, they still need improvements before being suitable for clinical use. The main downsides of these algorithms are the processing times, with accurate segmentations taking hours, or even days, to obtain. Additionally, both methods are not cleared by regulatory bodies (such as the FDA) for clinical practice.
Comparative research by Bartel et al. shows that FreeSurfer performs much better than both FSL-FIRST and manual segmentation, with resulting atrophy rates being predicted more accurately. Additionally, FreeSurfer showed good reproducibility through predicting similar atrophy rates using scan-rescan MRIs from the same patient. However, other methods have shown even better performance than FreeSurfer. Bartel et al. tested different available non-linear registration methods to predict atrophy rates. For such methods, an initial baseline segmentation is needed, which can be obtained either manually or automatically. The baseline segmentation is deformed to the follow up MRI image from which atrophy rates are predicted. See figure 3 for a visual explanation of registration-based segmentation methods. ANTs, Elastix, and NiftyReg are non-linear registration methods which were highly consistent in determining atrophy rates.2,3
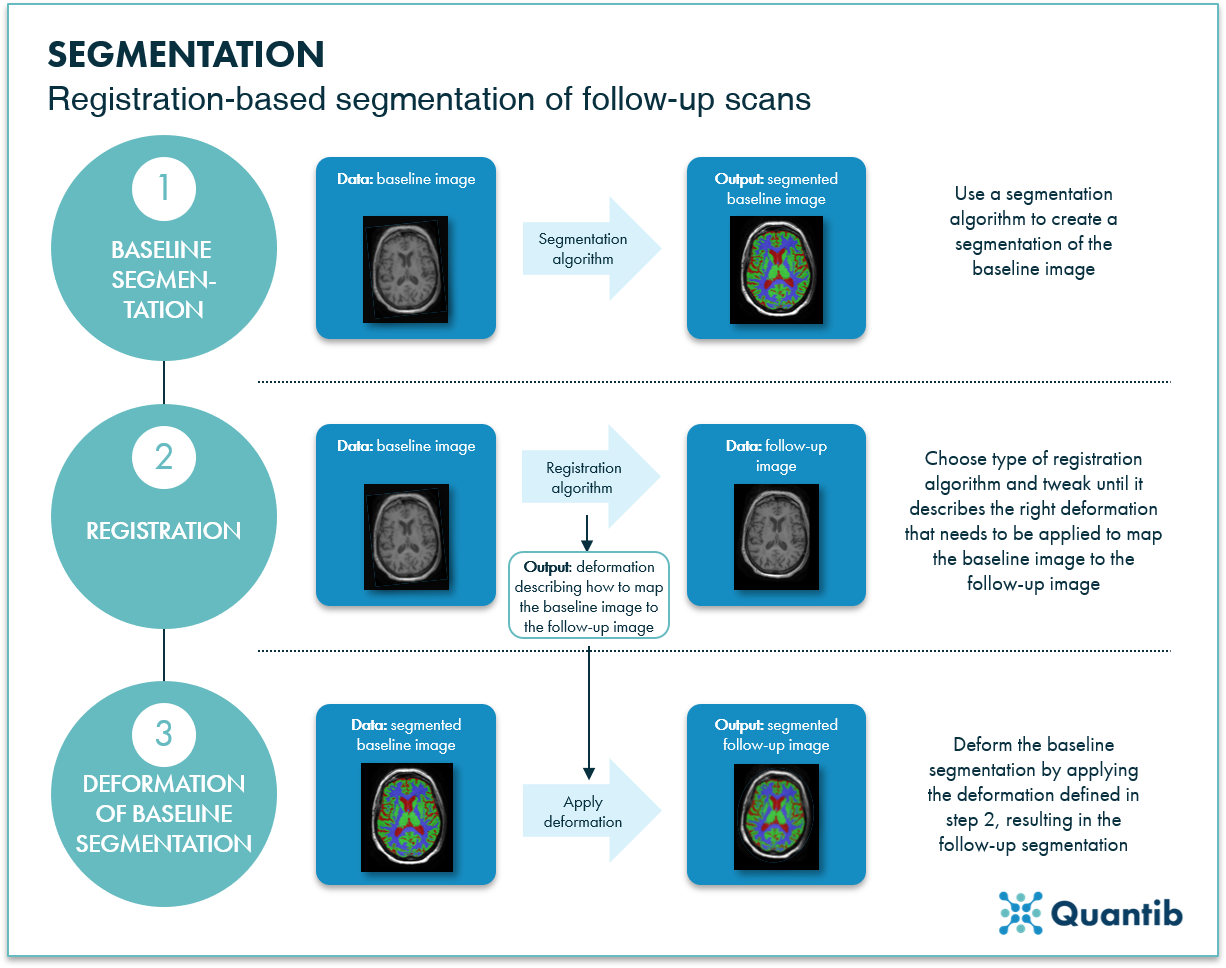
Figure 3: Segmentation of a follow-up image can be calculated by applying a registration algorithm to the segmentation of a baseline image.
Semi-automatic hippocampal volume segmentation
Additionally, Bartel et al. investigated a method called FASTSURF. FASTSURF is a semi-automatic segmentation method reducing manual segmentation time by requiring an observer to outline contours on only a few slices of an MR image, with the rest being computed automatically. As a result, segmentation time decreases by at least 30% without compromising segmentation quality.
Bringing automatic hippocampal volume segmentation to the clinic
To reiterate, accurate hippocampus segmentations can provide valuable diagnostic information. Although manual segmentation is simply too time consuming, and it results in substantial inter-observer variability, many automated segmentation solutions are currently not cleared for clinical usage yet.
To bring segmentation of the hippocampi to the clinic, Quantib developed an automated method which is included in an FDA cleared and CE marked radiology software product. Learn more about this product here.

