A few years ago the FDA cleared the first medical software using deep learning (DL) networks. This opened the door to many more companies applying for regulatory approval for their DL-based products. Amongst these products, many aim to classify medical images into certain categories, such as “this image may contain a malign tumor” versus “this image has a malign tumor”. DL is extremely suited to automate the solving of these classification problems. But which DL networks work best for these tasks? And what are the main characteristics of these networks?
In this article we take a deep dive on classification algorithms. We explore different network architectures, their characteristics and applications in the radiology field.
What is classification exactly?
Classification methods dived the input they are handed into groups or “classes”. For example, an algorithm which assesses a mammogram and will provide an answer to the question whether a patient has breast cancer, yes or no, is a classification algorithm. Additionally, any algorithm that a certain discreet scoring, such as BI-RADS, is just as much a classification algorithm.
Why use deep learning for classification in radiology?
Neural networks are very suited for classification. This has to do with the way the algorithm extracts features. In “old-fashioned” machine learning networks, features are designed manually. This means that the developer needs to sit down, dive in and figure out whether, for example, the feature average image intensity is important for determining if an image contains cancer.
Deep learning networks, on the other hand, ease this task by figuring out, based on the data, which features are important. The only thing you have to do is feed the data to the network (including the ground truths) and let the training procedure ‘teach’ the network (i.e. by updating its weights/parameters) how to best perform the task at hand.
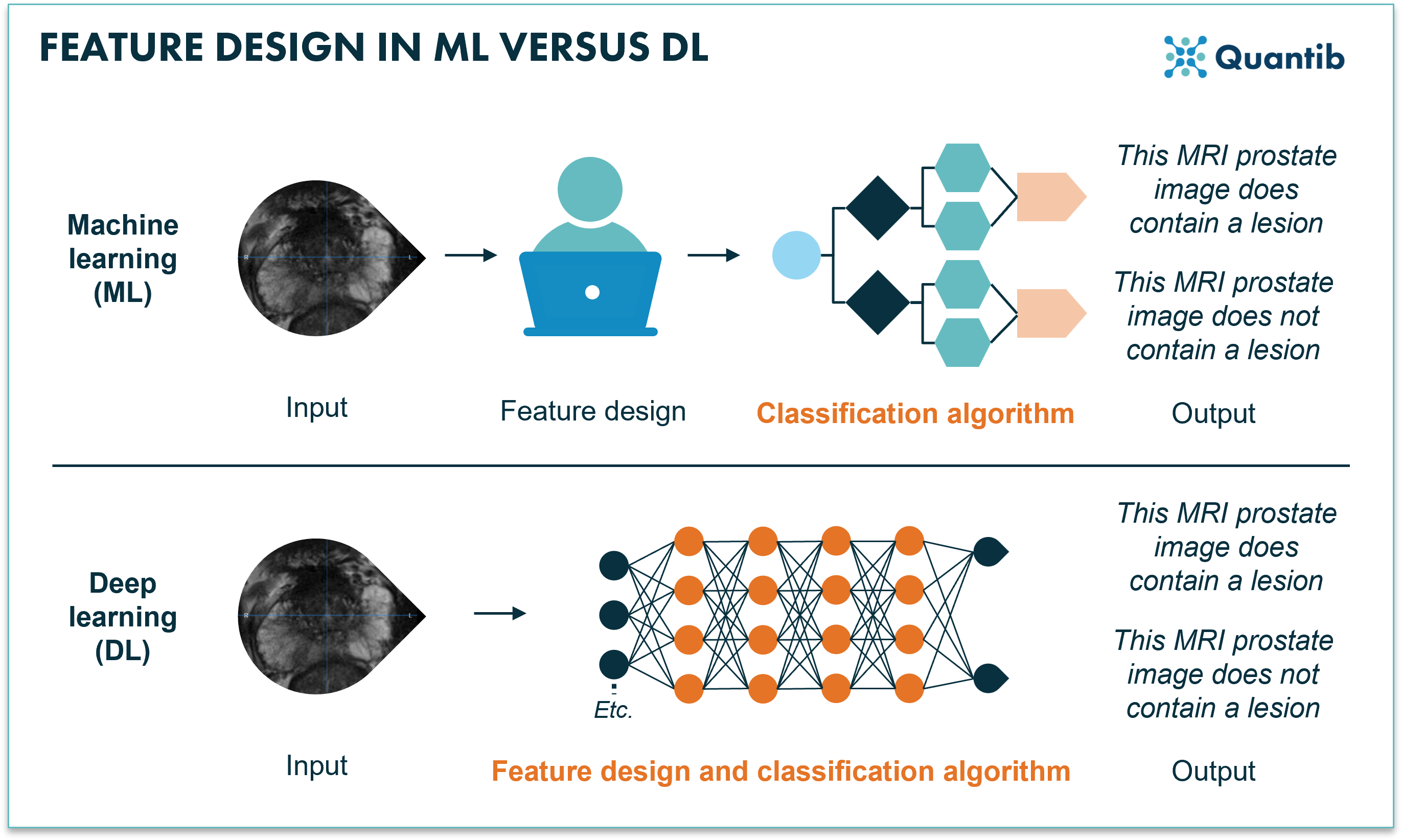 Figure 1: Machine learning requires manual feature designing before the training of the algorithm can start. Deep learning combines feature extraction and the training of the algorithm using a neural network.
Figure 1: Machine learning requires manual feature designing before the training of the algorithm can start. Deep learning combines feature extraction and the training of the algorithm using a neural network.
Which deep learning networks are the best for classification in radiology and how do they work?
Have you ever asked a vendor about the type of AI their software uses and gotten an answer you did not understand? Not anymore! Here we discuss some of the most used neural networks for classification tasks.
It all started with AlexNet
In 2012 Alex Krizhevsky competed in the ImageNet classification contest, the result: AlexNet. A deep convolutional neural network (CNN) with surprisingly good performance in classifying images into 1000 classes1. What was so special about this AlexNet? Mostly, the fact that it combined a whole lot of new techniques to the training of a CNN. For example, applying dropout, using multiple GPUs to cut back on training time, applying data augmentation and using a specific activation function which also dramatically decreased time needed to create the algorithm (for the ones interested in the details, we are talking about the ReLU fuction). All these techniques became pretty much standard in DL2.
AlexNet applied to radiology
Research of Hosny et al. shows an example of AlexNet applied to medical images. A network was developed to classify skin lesions into seven different classes. The strong performance metrics show the suitability of such an algorithm for classification tasks. Accuracy, sensitivity, specificity and precision all ranged between 95 and 100%3.
The next step: VGGs
Then VGG neural networks appeared, short for visual geometry group. The main goal of these networks was to increase the depth, i.e. the amount of layers of the neural network, as more layers would allow for addressing more details in the classification process. However, the deeper the network, the longer the training (and running!) time. So straight out of the box, these algorithms would provide results, but it would take a lot longer to provide you with an answer. What was the great knack applied in VGGs? They use very small kernels. This basically means that when taking information from the image and pushing this from one layer to the next, the network only combines information from a few voxels in the image that are right next to each other. AlexNet, on the other hand used larger kernels at the first stages of the network, which combine information from more voxels that are further apart4,5.
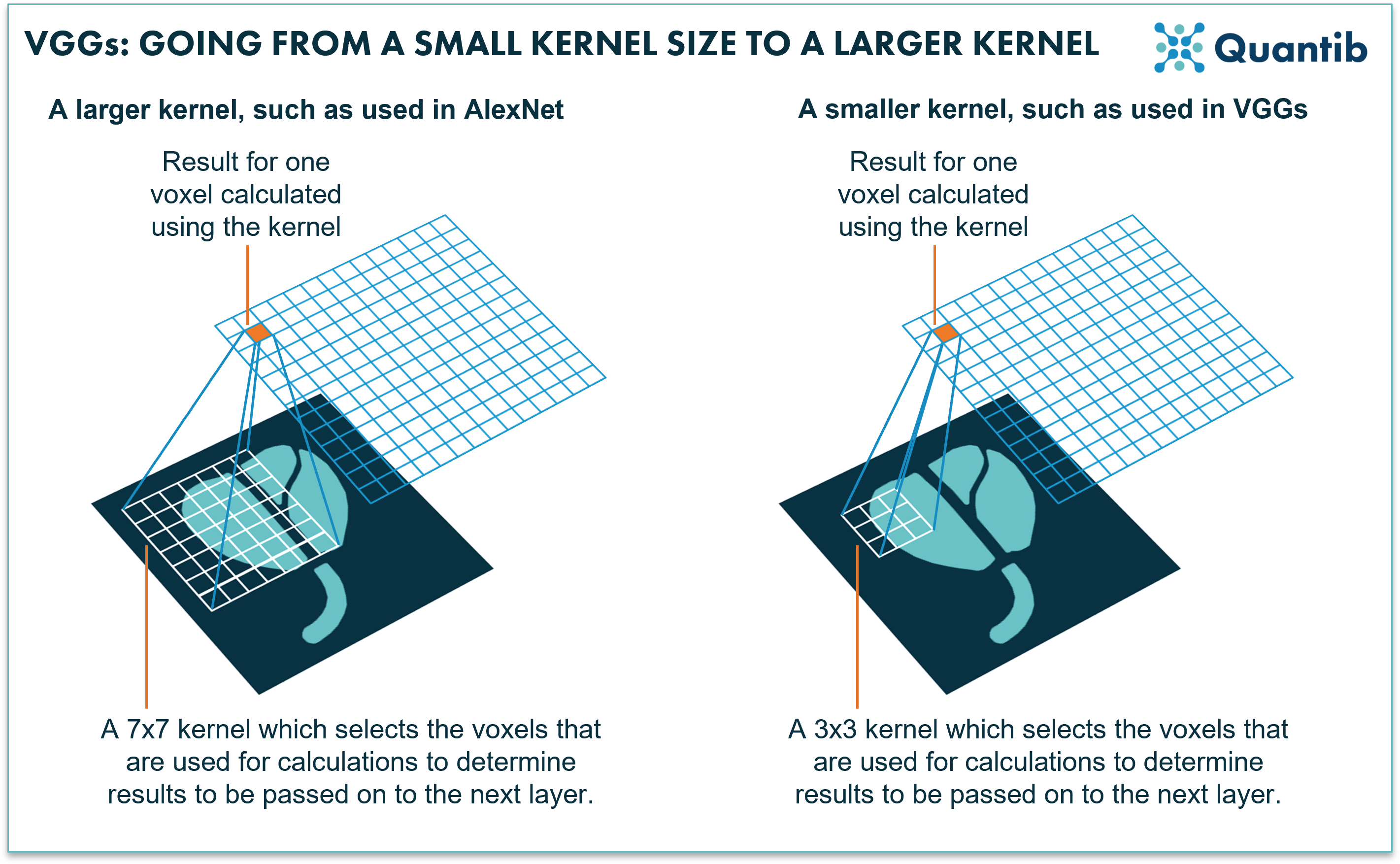 Figure 2: AlexNet uses relatively large kernels; rasters that move over an image to select voxels for calculations. VGGs use relatively small kernels.
Figure 2: AlexNet uses relatively large kernels; rasters that move over an image to select voxels for calculations. VGGs use relatively small kernels.
VGG applied to radiology
As an example, a recent study by Sitaula et al. focused on the computer-aided diagnosis of COVID-19 using a VGG model. A DL network reviews chest-X-rays (CXR) to detect the presence of COVID-19 suspect regions. Authors conclude that the algorithm performance suggests suitability of their method for CXR image classification of COVID-19 cases6.
Let’s keep on improving: the ResNet family
Unlikely the last step, but currently the most used algorithms in this same development path are those of the ResNet family. We stick to the design of CNNs, but yet another improvement is added, that of “identity shortcut connections” between the layers, also called “skip connection”. You can see these shortcuts as a back alley: they take information from a specific layer in the deep neural network and feed that directly into the final result or to a layer further down the network7. This enables each layer of the network to learn extra information compared to its input coming from the layer above. Additionally, skip connections simplify and accelerate the training.
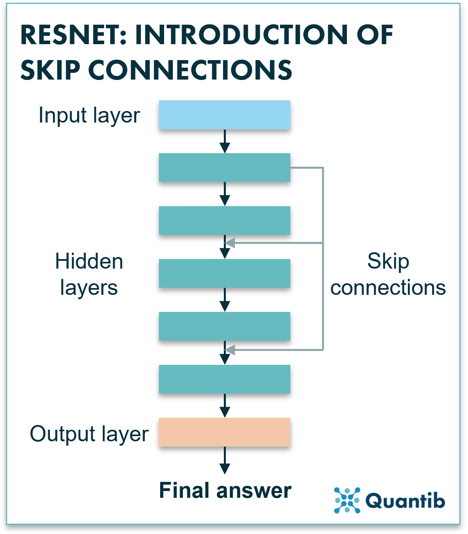 Figure 3: A ResNet uses skip connections to pass information from layers which are higher up in the network down to layers that are more to the bottom. This allows the network to avoid losing too much of the original input information which would make the algorithm diverge strongly.
Figure 3: A ResNet uses skip connections to pass information from layers which are higher up in the network down to layers that are more to the bottom. This allows the network to avoid losing too much of the original input information which would make the algorithm diverge strongly.
Within the ResNet family, ResNext is one of the newer versions of high performing CNNs. A ResNext actually combines multiple paths of stacks of hidden layers. Is this starting to sound a little abstract? Have a look at figure 4. Very simply said, you are copy-pasting a slight variation to the original ResNet and creating an expanded neural network by placing all these networks in parallel. Research shows that enlarging the network in this way actually leads to better performance than increasing the number of layers or the number of features in a network8,9.
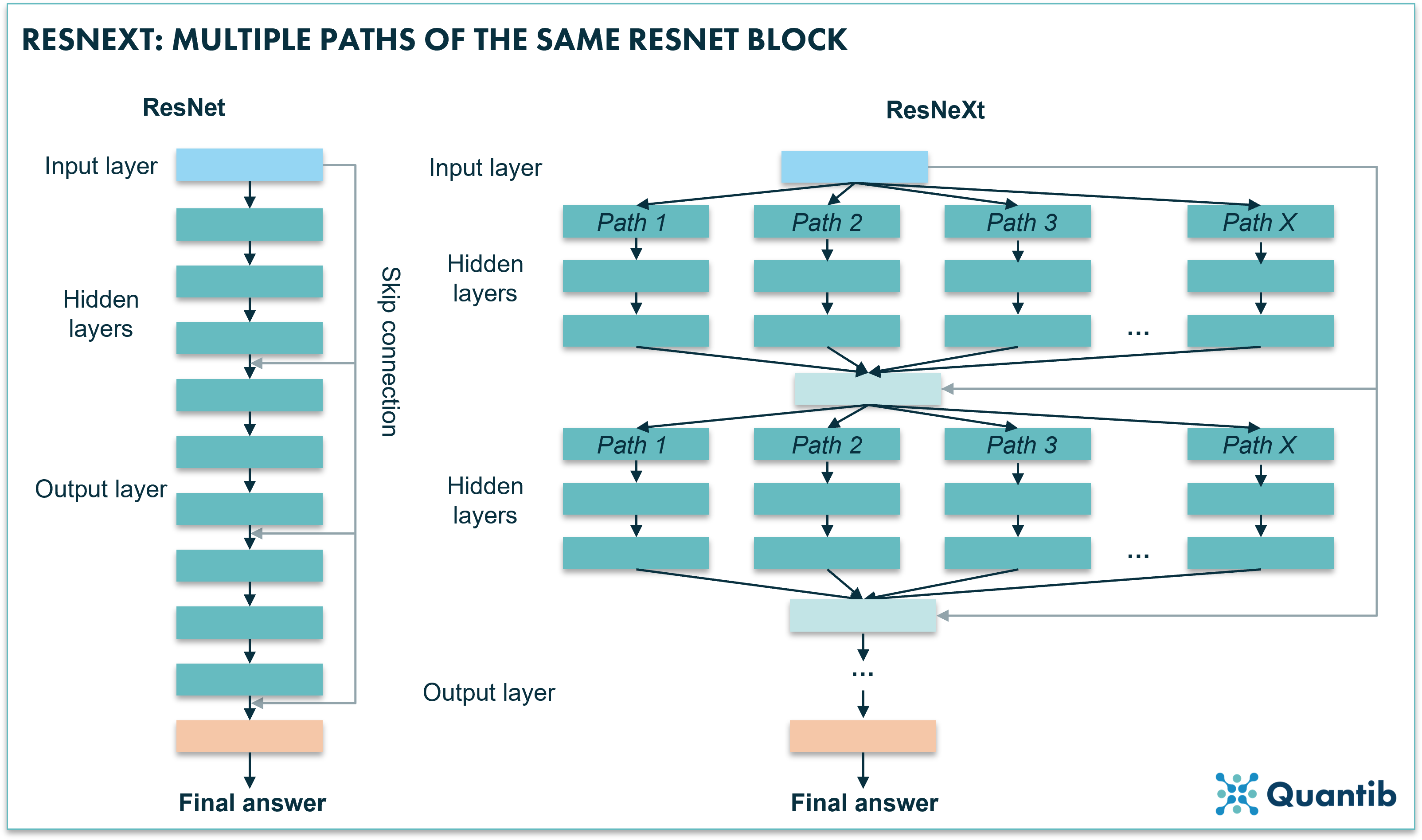 Figure 4: A ResNeXt uses multiple paths similar to a block of a ResNet algorithm. The algorithm feeds the input into these range of paths. Each path is trained in a slightly different way, which causes each path to learn slightly different things, hence, an opportunity for the network to learn more and therefore perform better.
Figure 4: A ResNeXt uses multiple paths similar to a block of a ResNet algorithm. The algorithm feeds the input into these range of paths. Each path is trained in a slightly different way, which causes each path to learn slightly different things, hence, an opportunity for the network to learn more and therefore perform better.
ResNet applied to radiology
Witowski et al. trained a ResNet to predict the probability of breast cancer in patients based on DCE-MRI. Performance checks on both an internal test set as well as on a retrospective study show good performance (area under the receiver operating characteristic curve of 92% and no significant difference between board certified-radiologists and the AI system, respectively). Additionally, the authors demonstrated generalizability on multiple datasets from different countries and different tumor types10.
A different approach: vision transformers
Vision transformers (ViTs) are fundamentally different to the deep neural networks discussed in the examples above. They do not use convolutional layers as CNNs do, but they use so-called transformer layers. Let’s dive in a bit deeper to understand the difference between convolutional layers and transformer layers and how the latter make use of patches.
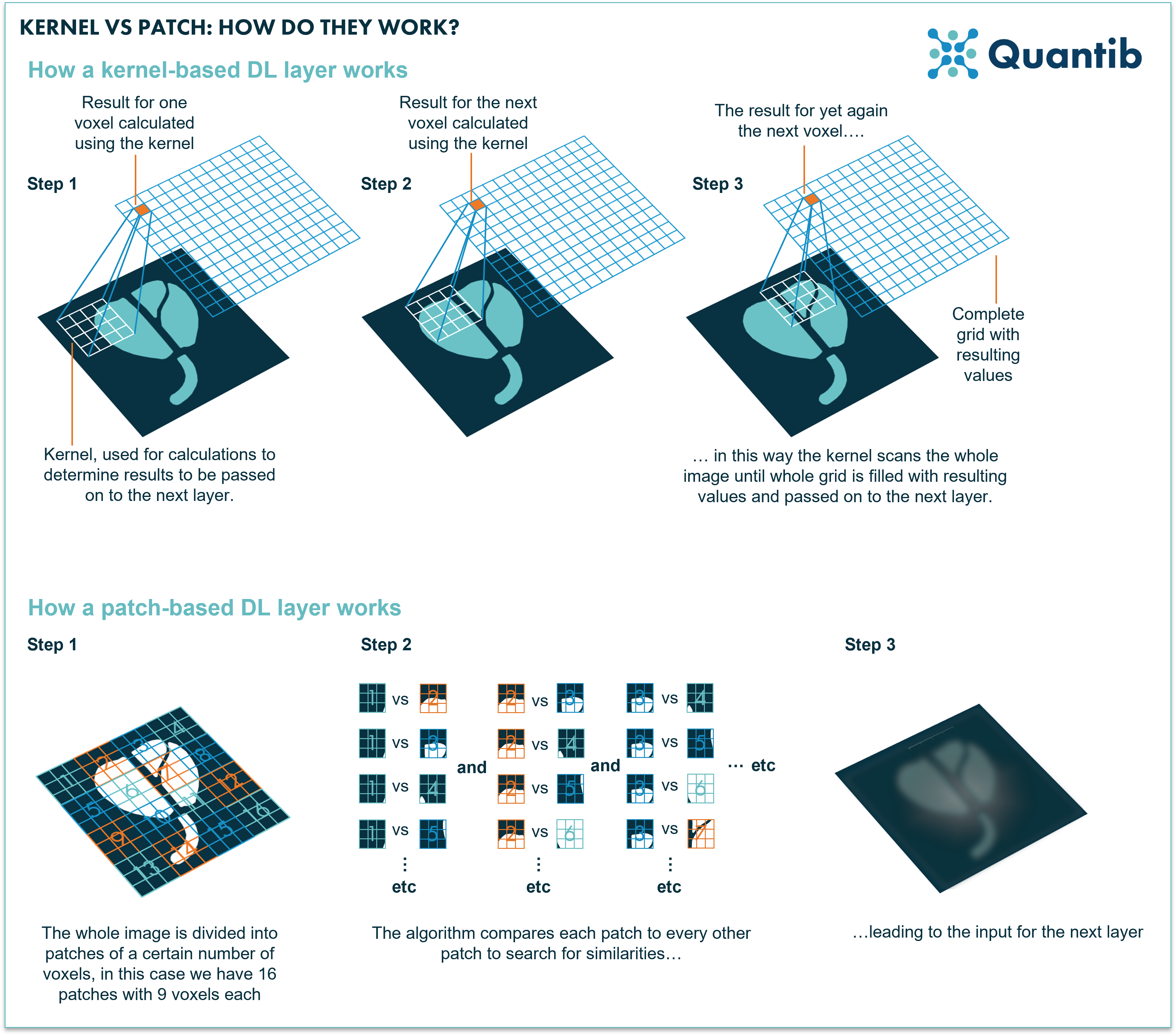
A convolutional layer uses kernels to assess images and derive information from them. A kernel is a rectangle-shaped set of pixels (actually most of the time a square). When the convolution is performed a kernel is applied to the entire input as a raster that moves across the image and takes as input the specific subset of adjacent voxels which are “chosen” by the kernel (see figure 5 for a visual explanation). This means that a convolutional layer performs the same operation independently of where it is located in the image. So if it is only starting in the upper left corner of the image it will perform the same calculation compared to when it is at the bottom right (with different input of course, namely the upper left set of voxels versus the bottom right). A convolutional layer uses several different kernels (which are also sometimes simply referred to as features) which are all learned together during training, i.e. the training of the network largely comes down to deciphering what these kernels should look like in order to get the algorithm to show the best performance possible.
Do you want to know more about Convolutional Neural Networks?
A transformer layer does things differently. The input to the layer is organized in patches, which are sets of voxels in a square. The transformer layer compares each patch to all the other patches in the image. Meaning it checks for relative importance of each set of voxels versus all the other same-sized sets of voxels throughout the complete image. This allows the algorithm to find correlations between image patches which were close to each other in the original image, but also between those that are at opposite sides of the input image. This means that the network could learn relevant relationships between parts of the image at any scale, which is generally not the case for convolutional networks. Hence, it can simultaneously tell you something about structures that cover a large part of the image, but also, for example, reveal a correlation between small lesions that can be found in multiple locations. Additionally, because a ViT network uses this comparison method and focuses on relations between different parts of the image, you are not constrained by using the same kernel for the whole image. This allows for more flexibility as well.
Altogether, a ViT provides a lot more freedom, however, it also requires vast amounts of computing power, making it too much of a stretch for current 3D medical images. Additionally, training of a vision transformer algorithm asks for significantly more data than other types of neural networks, also due to the increased degrees of freedom. Unfortunately, currently, such databases are hard to come by in medical imaging. The algorithms are also very suited though for natural language processing for which it is easier to gather (quality) data and training can happen in an unsupervised manner, hence no valuable time has to be spent on creating labels11.
Figure 5: A kernel selects a set of voxels which are relatively close to each other. These voxels are the input for calculation of the values that are passed on to the next layer. A transformer network divides the whole image in sets of voxels, so called patches. As a second step each of these patches is compared to all the other patches to check how similar these are. Lastly, this information is passed on to the next layer.
A PubMed search illustrates the challenges of applying ViTs to the field of medical imaging: only 22 results in the past year appear of which most are only distantly related to medical use cases and none of them apply the ViT to a 3D dataset. An interesting example, however, can be found in the work of Wang et al. who applied a ViT to the determination of the Kellgren-Lawrence scoring for osteoarthritis of the knee. According to the authors their classification method increases the accuracy by 2.5% compared to traditional CNN algorithms12.
The newest member of the family: ConvNext
Our last runner up: the ConvNext network. There is a reason why it is the last one to mention in this article, it is a combination of a ResNet and a ViT. The idea behind ConvNext was to ‘modernize’ the CNN architecture to implement the design hacks which made ViT the best performing method in computer vision classification challenges. These design choices included the most recent training techniques (e.g. the type of data augmentation that was used), and specific settings used in the network which make it more similar to a ViT, while still employing convolutional operations. Take the best of both worlds, so to say. For example, they revisited the use of larger kernel sizes to enable the network to see parts of the images that are further apart.
A PubMed search on ConvNext applied to classification problem illustrates the novelty of this technique, as it returns no results. Let’s hope that with advancing research and technology this promising method finds its way to medical image analysis as well.
How to measure the performance of classification networks in radiology?
Not every problem in medical imaging can be assessed in the same way. Some use cases require (automated) segmentations, some the answer to a classification question, others need localization of specific abnormalities. As the nature of these various types of assessments differs, they require different ways of performance measurement. What type of performance metrics work best in case of classification cases?
Since classification problems lead to a clear result per image that can be either wrong or right, testing an algorithm on a set of scans will lead to a certain number of true positives, true negatives, false positives and false negatives. Hence, metrics such as accuracy, sensitivity and specificity provide good context to evaluate a classification algorithm.
Curious to learn more about these metrics? Read our article on how to evaluate AI radiology algorithms.
Conclusion
Medical cases that require algorithms which can perform classification tasks are numerous. Therefore, a wide range of methods has been developed with this purpose, and research on how to optimize artificial intelligence methods for medical image processing is moving forward with grand strides. DNNs, such as AlexNet, VGGs and those of the ResNet family show vast results when applying them to diagnostic images.
Other promising, more recent developed algorithms, including ViT and ConvNext methods, have been successfully developed in computer vision. Unfortunately, they are still too premature to apply to medical images. Upon availability of more computing power and bigger, good quality datasets, however, there is great promise in the types of modern algorithms.
Bibliography
- Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet Classification with Deep Convolutional Neural Networks. http://code.google.com/p/cuda-convnet/ (2012).
- Wei, J. AlexNet: The Architecture that Challenged CNNs. Medium https://towardsdatascience.com/alexnet-the-architecture-that-challenged-cnns-e406d5297951 (2019).
- Hosny, K. M., Kassem, M. A. & Fouad, M. M. Classification of Skin Lesions into Seven Classes Using Transfer Learning with AlexNet. Journal of Digital Imaging 33, 1325–1334 (2020).
- Wei, J. VGG Neural Networks: The Next Step After AlexNet. Medium https://towardsdatascience.com/vgg-neural-networks-the-next-step-after-alexnet-3f91fa9ffe2c (2019).
- Boesch, G. VGG Very Deep Convolutional Networks (VGGNet) – What you need to know. Viso.ai https://viso.ai/deep-learning/vgg-very-deep-convolutional-networks/ (2021).
- Sitaula, C. & Hossain, M. B. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Applied Intelligence 51, 2850–2863 (2021).
- Feng, V. An Overview of ResNet and its Variants. Medium https://towardsdatascience.com/an-overview-of-resnet-and-its-variants-5281e2f56035 (2017).
- Xie, S., Girshick, R., Dollár, P., Tu, Z. & He, K. Aggregated Residual Transformations for Deep Neural Networks. (2016).
- Ma, E. Enhancing ResNet to ResNeXt for image classification. Medium https://medium.com/dataseries/enhancing-resnet-to-resnext-for-image-classification-3449f62a774c (2020).
- Witowski, J. et al. Improving breast cancer diagnostics with artificial intelligence for MRI. (2022) doi:10.1101/2022.02.07.22270518.
- Boesch, G. Vision Transformers (ViT) in Image Recognition – 2022 Guide. Viso.ai https://viso.ai/deep-learning/vision-transformer-vit/ (2022).
- Wang, Y., Wang, X., Gao, T., Du, L. & Liu, W. An Automatic Knee Osteoarthritis Diagnosis Method Based on Deep Learning: Data from the Osteoarthritis Initiative. Journal of Healthcare Engineering 2021, 1–10 (2021).